About YuanST Lab
Welcome to the YuanST (Spatial-Temporal) Lab at the Institute of Science and Technology for Brain-inspired Intelligence (ISTBI), Fudan University. We develop computational methods and spatial omics technologies to understand complex biological systems through multimodal data integration and spatiotemporal analysis. We focus on three interconnected areas: optimizing spatial omics experimental design through computational approaches, developing computational methods for integrating multimodal spatiotemporal data to represent tissue microenvironment, and mining these spatiotemporal data to extract meaningful biological insights and biomedical applications.
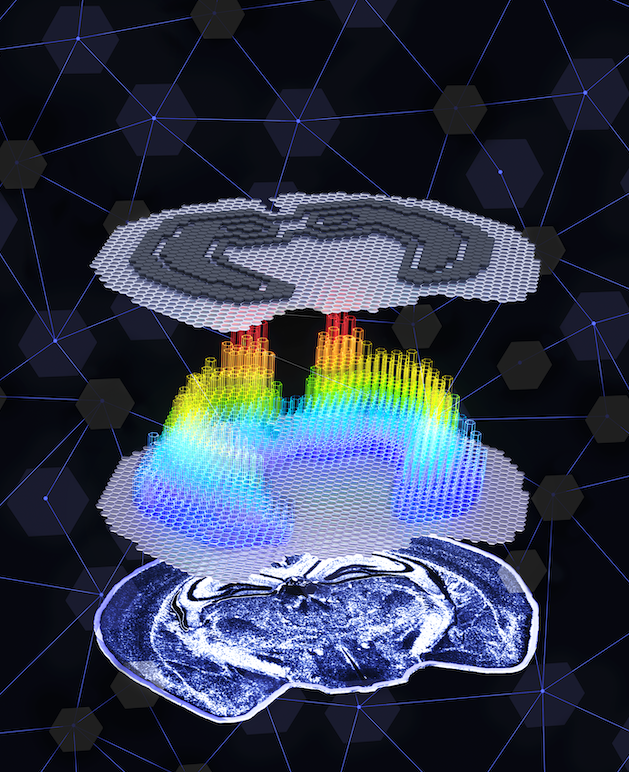
RESEARCH FOCUS
Optimization of Spatial Omics Experimental Design
Our lab is dedicated to developing innovative computational approaches to optimize spatial omics experimental design. We employ advanced algorithms and mathematical models to enhance the efficiency and accuracy of spatial transcriptomics and other spatial omics technologies. By carefully considering factors such as tissue architecture, cellular heterogeneity, and technical constraints, we design optimal sampling strategies and experimental protocols. Our computational frameworks help researchers maximize the information gained from each experiment while minimizing costs and technical variations. This work is fundamental to advancing the field of spatial omics and ensuring high-quality data generation for downstream analyses.
Computational Methods for Multimodal Spatiotemporal Integration
A core focus of our research is the development of sophisticated computational methods for integrating diverse spatiotemporal data types. We create advanced algorithms and tools to combine multiple layers of biological information, including spatial transcriptomics, proteomics, and imaging data, while preserving their spatial and temporal contexts. Our methods address the unique challenges of data alignment, normalization, and integration across different modalities and scales. We aim to construct comprehensive representations of tissue microenvironments that capture both spatial organization and temporal dynamics. These integrated approaches provide a more complete understanding of cellular interactions and tissue architecture than any single data type alone.
Mining Spatiotemporal Data for Biological Insights
The third pillar of our research focuses on extracting meaningful biological insights and biomedical applications from integrated spatiotemporal data. We develop and apply advanced machine learning and statistical methods to analyze complex spatial patterns, cellular interactions, and temporal dynamics within tissues. Our approaches help identify key biological mechanisms, disease markers, and therapeutic targets. We particularly emphasize the translation of our findings into practical biomedical applications, such as improving disease diagnosis, understanding development and aging processes, and identifying new therapeutic strategies. This work bridges the gap between computational analysis and clinical applications, contributing to advances in precision medicine and therapeutic development.
PRINCIPAL INVESTIGATOR

Zhiyuan Yuan (原致远)
Associate Professor at Fudan University
zhiyuan@fudan.edu.cn
Dr. Zhiyuan Yuan is a Associate Professor and Ph.D. supervisor at the Institute of Science and Technology for Brain-inspired Intelligence (ISTBI) at Fudan University, where he also serves as Deputy Director of the Center for Integrative Spatial-Omics Research. After receiving his bachelor's from Wuhan University and Ph.D. from Tsinghua University (advisor: Prof. Michael Q. Zhang), he was exceptionally recruited as an independent PI at Fudan in 2022 and was fast-tracked to his current position in 2025. His research focuses on genomic large language models and generative AI for single-cell and spatial omics. Dr. Yuan is a recipient of China's national high-level youth talent program and numerous other awards, with first or corresponding author publications in top journals such as Nature Methods, Nature Machine Intelligence, and Nature Communications. He actively leads national and municipal research grants, mentors students who also publish in high-impact journals, and serves on the youth editorial boards and committees of several professional organizations and journals.
原致远,就职于复旦大学类脑智能科学与技术研究院,长聘副教授/研究员,博士生导师,空间组学整合研究中心副主任。本科毕业于武汉大学计算机学院,博士毕业于清华大学自动化系,博士毕业后被破格引进复旦大学任独立PI(2022.9),后破格晋升为长聘副教授/研究员(2025.4)。
入选国家高层次人才特殊支持计划青年项目(2024),上海市各项人才计划(青年东方英才、晨光学者、扬帆计划、明珠菁英人才、上海35U35、启源青年学者)及复旦大学卓学人才计划等。第一/通讯作者身份获得世界人工智能大会青年优秀论文奖(10项/年,2023)、世界人工智能与生物医药十大进展(10项/年,2025)、中国生物信息学十大进展(10项/年,2022/2024)、周传纪念奖(10项/年,2023)等奖励。
担任腾飞书院及大数据学院本科生班导师。本科生指导成绩包括:取得国自然本科生项目立项;指导本科生一作发表于Nature Machine Intelligence(复旦在该期刊首篇本科生一作research article);学生获复旦大学本科生学术创新追求卓越奖学金(复旦大学本科生学术最高荣誉)、复旦大学优秀学生标兵、智擎学者、莙政学者、复旦大学奖学金获奖学生优秀典型、国家奖学金等奖励。研究生指导成绩包括:博士生取得中国科协青年科技人才培育工程博士生专项计划立项;学生获研究生国家奖学金;指导研究生一作工作发表于Nature Methods、Nature Protocols、Cell Genomics、Nature Communications等顶级期刊。
主持国家自然科学基金面上、青年项目、上海市计算生物学重点专项、曹娥江基础创新培育项目;连续3年主持腾讯AI Lab/CCF犀牛鸟专项研究基金(2023/2024/2025),并获得年度“优秀奖”;骨干参与国家重点研发计划、上海市重点专项等重大科研任务。
研究方向为AI+空间组学,建立系列人工智能算法解析细胞状态、行为、集群和动态变化。学术成果以第一或通讯作者(含共同)发表在 Nature Methods(2021/2023/2024/2025a/2025b)、 Nature Machine Intelligence(2025)、 Nature Protocols(2024)、 Cell Genomics(2024)、Nature Communications(2022/2024/2025a/2025b)等期刊。部分研究成果被权威期刊和国际同行正面评价:多次被选为Nature Methods、Nature Communications、Cell Genomics亮点论文(Featured Article)、Nature Methods头版新闻(News & Views),被Nature Methods研究简报(Research Briefing)特别报道,被Nature Biotechnology高亮推介(NBT Highlight),入选Nature Methods关注度排名前1%论文(Nature出版社官方统计),入选ESI高被引论文等。同时,代表性研究成果也被人民日报、新华社、环球网、复旦大学、上观新闻等官方媒体专门报道。
担任 Genome Biology (中科院1区) Editorial Board Member,Genom. Proteom. Bioinf.(GPB)、J. Genet. Genom.(JGG)青年编委。担任方法学(Nat. Methods/Nat. Biotechnol./Nat. Mach. Intell./Nat. Comput. Sci.),生物学(Nat. Genet., Nat. Metab., Nat. Struct. Mol. Biol., Cell Genom., Cell Syst.)及综合性(Nat. Commun., Science Advances, Natl Sci Rev, Cell Rep.)等>40种权威期刊审稿人。担任国际计算生物学学会中国理事会理事、中国自动化学会智能健康与生物信息专委会委员、中国计算机学会生物信息专委会委员、中国人工智能学会生物信息学与人工生命专委会委员、中国生物信息学会(筹)生物信息学算法研究专委会委员、上海市生物信息学会委员。
MEMBERS
We are actively looking for postdoc, graduate/undergraduate students, research assistants! Please find the information here.
Post-doctoral Associate

Daoliang Zhang (张道良)
2025 - Now
Education: PhD, Shandong University
GigaScience ‘24, Bioinformatics Advances ’25
Students

Fangyuan Zhao (赵方圆)
2022 - Now
Joint Ph.D. Student
Education: PhD Candidate, Institute of Computing Technology,CAS
Nature Methods ’24
Nature communications ’25

Zhikang Wang (王智康)
2024 - Now
Research Assistant
Education: PhD Candidate, Monash University
Nature Machine Intelligence ’25
Nature Methods ’25
Nature Methods ’25

Qi Zou (邹麒)
2024 - Now
Joint Ph.D. Student
Education: PhD Candidate, Shandong University
Nature Methods ’25

Chuangyi Han (韩创意)
2024 - Now
Undergraduate
Education: BSc Candidate, Fudan University
Nature Machine Intelligence ’25
Nature Methods ’25

Kairan Kang (康恺然)
2024 - Now
Ph.D. Student
Education: BSc, Tongji University

Yixuan Du (杜宜轩)
2024 - Now
Ph.D. Student
Education: BSc, Tongji University

Tianyi Gu (顾添奕)
2024 - Now
Joint Ph.D. Student
Education: PhD Candidate, Fudan University

Yunzhi Yan (严筠芷)
2024 - Now
Joint Ph.D. Student
Education: PhD Candidate, Fudan University

Chengcheng Sun (孙成成)
2024 - Now
Joint Ph.D. Student
Education: PhD Candidate, Fudan University

Yinghao Zhang (张英豪)
2024 - Now
Joint Ph.D. Student
Education: PhD Candidate, Fudan University

Xinwang Yang (杨鑫旺)
2024 - Now
Joint Ph.D. Student
Education: PhD Candidate, Harbin Institute of Technology University

Han Shu (舒涵)
2024 - Now
Joint Ph.D. Student
Education: PhD Candidate, Northwestern Polytechnical University
The Innovation ’25

Sijie Li (李思婕)
2025 - Now
Ph.D. Student
Education: PhD Candidate, Fudan University

Jiabei Cheng (程佳贝)
2025 - Now
Joint Ph.D. Student
Education: PhD Candidate, Shanghai Jiao Tong University

Wenyang Liu (刘文扬)
2025 - Now
Ph.D. Student
Education: BEng, Zhejiang University

Zheqi Hu (胡哲琪)
2025 - Now
Joint Master Student
Education: Master student, Harbin Institute of Technology University (ShenZhen)

Xiangming Yan (严翔明)
2025 - Now
Joint Ph.D. Student
Education: Ph.D. student, Harbin Institute of Technology University (ShenZhen)
Alumni

Yan Cui (崔岩)
2023 - 2024
Research Assistant
Nature Communications ’25
Nature Methods ’25
Current position: PhD student, University of Pennsylvania

Na Yu (于娜)
2024 - 2025
Joint Ph.D. Student
GigaScience ‘24
Current position: Post-doctoral Associate, Westlake University

Tong Pan (潘彤)
2025 - 2025
Visiting Student
Current position: Postdoc research fellow, Adelaide University
PUBLICATIONS
*: These authors contributed equally to this study and share the first authorship.
#: Co-corresponding author
TALKS
Invited Talks
2025.12.18 上海
多组学质量控制与数据整合高峰论坛 “空间组学数据整合”
2025.12.05 西安
第七届全国生物医学数据挖掘与计算学术会议 “时空组学的智能计算与生物解析”
2025.11.20 上海
第八届上海国际肿瘤内科学大会 “空间组学数据整合”
2025.11.14 厦门
“定量生物学:数学建模与统计推断”研讨会通知 “空间组学数据的智能分析与挖掘”
2025.11.07 上海
上海市生物信息年会 “计算空间组学”
2025.10.26 杭州
the 20th Annual Meeting of the International Conference on Genomics “High-Content 3D Digital Organ Construction”
2025.10.07 南宁
第十四届全国生物信息学与系统生物学学术大会 “融合空间信息的跨组学翻译方法”
2025.08.08 海口
第十届中国计算机学会生物信息学会 “空间多组学计算方法进展” 邀请报告
2025.07.22 长沙
中国解剖学会组织学与胚胎学分会2025年学术年会 “人工智能赋能的空间多组学” 分会场邀请报告
2025.07.19 东莞
第三届松山湖数学论坛 “人工智能赋能的单细胞时空分析” 分会场邀请报告
2025.06.26 北京
生命科学前沿技术发展战略对策研讨会 “空间多组学数据整合” 分会场邀请报告
2025.06.25 上海
上海交通大学转化医学研究院 “空间多组学数据整合” 邀请报告
2025.05.23 济南
第十一届全国计算生物学与生物信息学学术会议 “空间多组学数据整合” 分会场邀请报告
2025.05.10 北京
第12届“数学、计算机与生命科学交叉研究”青年学者论坛 “计算空间组学” 主会场邀请报告
2025.04.25 上海
第三届 CCF 生物信息学“新未来”青年学者研讨会 “计算空间组学” 邀请报告
2025.04.12 武汉
武汉大学人工智能学院 “计算空间组学” 邀请报告
2025.03.29 深圳
第二届全国基因组信息学大会 “计算空间组学” 分会场邀请报告
2025.01.20 上海
上海交通大学溥渊未来技术学院 “空间组生物信息学” 邀请报告
2025.01.17 上海
“思享前沿”学术研讨会暨2024年上海市计算机学会生物信息学专委会年会 “空间组生物信息学” 邀请报告
2024.12.15 上海
The 2nd Med-X International Conference “AI-aided spatial omics experimental design” 分会场邀请报告
2024.12.06 广州
第六届全国生物医学数据挖掘与计算学术会议 “空间组学数据的智能分析与挖掘” 分会场邀请报告
2024.11.29 沈阳
第二届CCF数字医学大会 “AI辅助空间组学实验设计” 分会场邀请报告
2024.11.27 北京
第三届中国医学基础学科发展大会 “AI辅助空间组学实验设计” 分会场邀请报告
2024.11.23 杭州
第二届人工智能与生物信息学前沿交叉青年论坛 “空间组生物信息学” 邀请报告
2024.10.26 上海
第二十届上海大肠癌论坛 "空间组学解析组织微环境" 大会报告
2024.10.12 海口
第十三届全国生物信息学与系统生物学学术大会 "AI辅助空间组学实验设计" 大会报告
2024.09.14 济南
山东大学 "空间组学生物信息" 邀请报告
2024.08.09 长春
第九届中国计算机学会生物信息学会议 "空间组学生物信息" 分会场邀请报告
2024.06.06 上海
同济大学 "空间组生物信息学解析组织微环境" 邀请报告
2024.05.24 青岛
第二届山东省生物信息学学术大会暨中国生物信息学学会(筹)生物信息学算法研究专委会学术大会 "空间组生物信息学解析组织微环境" 分会场邀请报告
2024.04.18 福州
第三届中国智能健康与生物信息大会 "空间组生物信息学解析组织微环境" 分会场邀请报告
2024.01.19 上海
复旦大学影像和空间组学论坛 "空间生物信息学" 邀请报告
2023.11.14 上海
复旦大学 "Modeling spatial and temporal omics data" 邀请报告
2023.11.08 上海
计算神经科学与类脑智能教育部重点实验室2023年度学术委员会 "空间生物信息学" 邀请报告
2023.11.06 北京
北京大学 "Exploring Tissue Architectures with Computational Spatial Omics" 邀请报告
2023.11.05 北京
中国生物工程学会第十五届学术年会暨2023年全国生物技术大会 "空间生物信息学" 分会场邀请报告
2023.11.03 上海
第四届国际人类表型组研讨会 "Dissecting Tissue Microenvironment with Spatial Bioinformatics" 分会场邀请报告
2023.10.20 无锡
第三届数学生命科学大会 "空间生物信息学解析组织微环境" 分会场邀请报告
2023.08.26 三亚
崖州湾国家实验室 "Exploring Tissue Architectures with Computational Spatial Omics" 邀请报告
2023.08.16 上海
中国科学院营养与健康研究所 "用计算空间组学探索组织结构" 邀请报告
2023.06.09 杭州
之江实验室 "Exploring Tissue Architectures with Computational Spatial Omics" 邀请报告
2023.05.26 杭州
浙江工业大学信息学院 "Exploring Tissue Architectures with Computational Spatial Omics" 邀请报告
2023.05.20 上海
复旦大学庆学术报告“Exploring Tissue Architectures with Computational Spatial Omics”邀请报告
2023.04.20 上海
复旦大学影像和空间组学论坛 "空间组学探索组织微环境" 邀请报告
2023.04.15 线上
"基因组与AI"一作面对面论坛 "SODB facilitates comprehensive exploration of spatial omics data" 邀请报告
2023.04.12 上海
复旦大学生命科学学院 "空间组学数据中微环境的建模与分析方法" 邀请报告
HONORS & AWARDS
-
1.
韩创意,2025年国家自然科学基金青年学生项目(国家级,全校仅20人)
-
2.
韩创意,2025年国家奖学金(国家级)
-
3.
韩创意,2025年复旦大学智擎学者(校级)
-
4.
韩创意,2025年复旦大学莙政学者(校级)
-
5.
韩创意,2025年复旦大学优秀学生标兵(校级)
-
6.
李思婕,入选2025年度中国科协青年科技人才培育工程博士生专项计划(国家级)
-
7.
张道良,2025年复旦大学"超级博士后"(校级)
JOIN US
直博生
我们每年在保研夏令营中招生,在夏令营官方考察之前,需在课题组提前实习(夏令营当年4月之前【投递简历】,之后【课题组内面试】、【进组考察】)。要点:
【投递简历】
建议您是信息学或数学相关背景,或是有较强深度学习基础的生物背景学生。请在简历中突出真实科研经历。
【课题组内面试】
通过简历筛选后,需面向课题组内成员讲解您的本科学习、科研、竞赛经历,及未来科研规划。面试过程中您也可以向组内成员提出问题,达到双向了解和选择目的。
【进组考察】
面试通过后,课题组长会布置1个测试题目,要求您在课题组内线下/线上实习期间内(约3-5周)完成。
普博生
课题组也招收申请考核制博士生(名额取决于夏令营招生情况),在官方申请考核之前,需在课题组提前实习(拿到硕士学位前一年6月30日之前【投递简历,需包含硕士导师的推荐信一封】,之后【课题组内面试】、【进组考察】)。要点:
【投递简历】
建议您是信息学或数学相关背景,或是有较强深度学习基础的生物背景学生。请在简历中突出真实科研经历及论文列表。且需要硕士导师的推荐信一封。
【课题组内面试】
通过简历筛选后,需面向课题组内成员讲解您的学习、科研经历,及未来科研规划。面试过程中您也可以向组内成员提出问题,达到双向了解和选择目的。
【进组考察】
面试通过后,课题组长会布置1个测试题目,要求您在课题组内线下/线上实习期间内(约3-5周)完成。